Undergraduate education-Michigan State
Graduate education-Loyola University
Post-Doc experience-Northwestern University
Research Scientist experience-University of Chicago
Coursework-MSU and Loyola University
Born in Michigan, I grew up in the north suburbs of Detroit. I went to highschool at Notre Dame Highschool in East Pointe. There I gained my interest in the sciences.
After graduation, I enrolled in Lyman Briggs College at Michigan State University. Lyman Briggs College is a residential learning community which focused on core scientific disciplines: physics, chemistry, and biology. In addition, it balanced out the coursework with ethics and sociology courses focusing on the relationship between science and impact on society. I found the environment stimulating because it not only rigorously trained me in the core science, but also taught me an appreciation for the implications of science on society. After my core coursework, I chose a major in microbiology and took courses in bacterial pathogenesis, bacterial genetics, eukaryotic biology and virology. To obtain laboratory experience, I worked as a lab technician in the laboratory of Dr. Norbert Kaminski in the Department of Pharmacology and Toxicology. The focus of the laboratory was on immunotoxicology and the effects of the dioxin and cannabinoid receptors on immune cell function. During this time, I got to interact with research staff and graduate students. I enjoyed working in a development and research environment and decided to further my education in graduate school.
I was accepted into the Molecular Biology Program at Loyola University in Maywood, Illinois. The program focused on training in molecular biology techniques and their application to cancer biology, cell signaling, wound healing, bacteriology and virology projects. From my previous coursework at Michigan State, I had developed an interest in virology. Viruses propagate through production of protein/lipid capsules containing the necessary genetic information to produce more progeny. When presented by obstacles to replication, they have the ability to adapt in unique and often unexpected ways. This aspect intrigued me and lead me to pursue a virology research project.
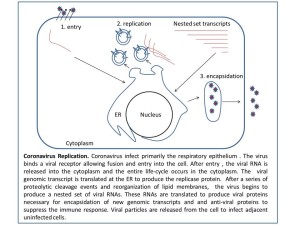
In 2002, I joined the laboratory of Dr. Susan Baker PhD. The laboratory focuses on Coronavirus replication, more specifically papain proteases and their involvement in directing replicase assembly. Coronavirus are enveloped positive strand RNA viruses. Coronaviruses infect most mammals and birds including humans, birds, bats, civeats, and mice. Each species usually has a unique variant of the family found in the population IBV (birds), MHV (mice), HCov (humans). Coronaviruses infection cause encephalitis, gastro-intestinal and upper respiratory distress. Before 2003, there were no recorded accounts of coronaviruses causing severe disease in humans. In 2003, in Guangdong province a virus was isolated from patients with severe acute respiratory distress, the virus was later identified as a coronavirus (sever acute respiratory syndrome virus, SARS-CoV) and found to be the etiological agent of the disease. Later a second coronavirus (NL63) was identified in the Netherlands and shown to be one of the contributing viruses in causing Crop in children. More recently in 2012, a third coronavirus has been isolated in the middle east (MERS-Cov) and shown to be causing upper respiratory distress.
The lab studies the production of the replicase proteins which form complexes on membrane vesicles. After entry of the virus, the genomic RNA is translated to produce one long inactive polymerase polyprotein. In addition to being very long, the polyprotein is synthesized as a multi-pass integral membrane protein associated with the ER. A series of 2-3 coronavirus proteases (cysteine and papain) cleave the polyprotein at specific cleavage sites. The small protein subunits then associate with each other and redirect membranes into vesicles. These vesicle membrane complexes become the site active site of virus replication. The genomic RNA is fed through the replicase complex to produce new copies of the genome. In Susan’s lab, I learned about protease function, replicase assembly and inhibitors of protease function. I cloned the SARS-Cov segments of the cysteine protease and demonstrated auto-cleavage of the polyprotein. I was also involved in the initial attempts to develop a reporter construct for screening protease inhibitors. I produced viral recombinant proteins and injected them into rabbits to produce polyclonal antibodies used for the detection of viral proteins during infection.
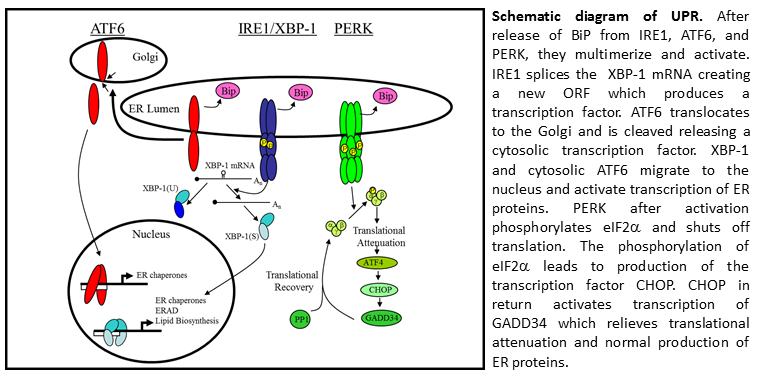
My main project involved studying the activation of the host stress responses during viral replication. Since the virus produce large amounts of ER proteins and rearranges the host ER, we suspected the virus activated the host stress response pathway called the Unfolded Protein Response (UPR). This project was done in collaboration with an UPR expert previously in the department, Dr. Joseph Brewer. The UPR is a stress response pathway utilized by cells to slow protein production, enlarge the ER, and enhance protein production and quality control proteins. In B cells, ER expansion is required for B cell differentiation into a plasma cell. Since plasma cells produce large quantities of antibodies (a multi-meric glycosylated protein with cysteine bonds) the ER needs to expand several times and produce proteins capable of antibody folding, monitoring antibody glycosylation, and enhancing cysteine bond formation between antibody chains.
My main project involved examining the three arms of the UPR and determining their level of activation during infection. I found the cell had activated the UPR response signal transducers (ATF6 and IRE1) but did not produce a transcriptional response. I found evidence infected cells initiated translation repression through eIF2a phosphorylation. The repression was not relieved because viral infection inhibited the production of GADD34, a phosphatase responsible for dephosphorylating eIF2a and reinitiating translation. (Journal of Virology, May 2008, vol. 82 no. 9 4492-4501).
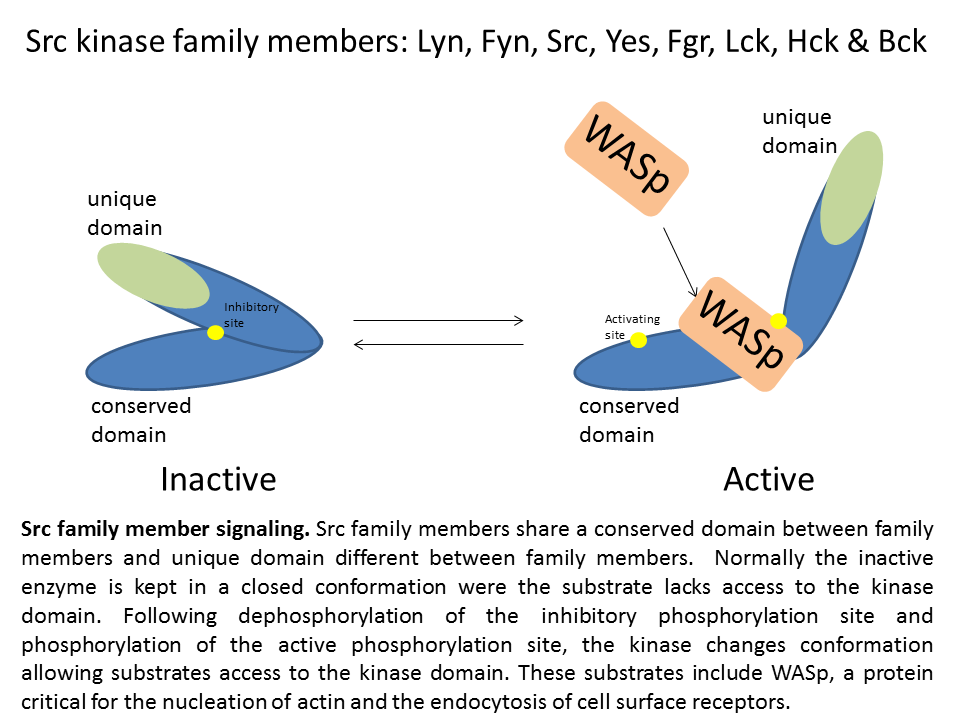 After my graduate work, I wanted to get experience in a different area of research. I therefore, joined the laboratory Dr. SJ Corey MD at Northwestern University. The laboratory studies hematology and oncology disorders. The laboratory focuses on receptor tyrosine kinases and src kinase associated signaling. Src kinases phosphorylate a variety of targets and are direct cellular processes such as endocytosis of receptors, cell proliferation, cell migration metabolism and differentiation. Since Src kinases are implicated in a wide variety of diseases, they have become a target of pharmacological treatment. Inhibitors such as dasatinib (Bristol-Myer Squibb) bind to the active kinase domain and block the function of the Src kinase, Lyn, and the kinase domain of the fusion protein bcr-abl. Other Src kinase/bcr-abl have entered into clinical trials for tumors, including saracatinib, bosutinib, and KX01 (kinex pharmaceuticals). In addition to Src, the Src family contains several other family members: Lyn, Fyn, Yes, Fgr, Hck, Lck, and Bck. The family members share the same kinase domain but each have their own unique domain. Each family member has its own cell type distribution. Src kinases Lyn, Hck, and Fyn are distributed in immune cell types and regulate the function of inflammatory responses by T cells, B cells, and macrophages.
After my graduate work, I wanted to get experience in a different area of research. I therefore, joined the laboratory Dr. SJ Corey MD at Northwestern University. The laboratory studies hematology and oncology disorders. The laboratory focuses on receptor tyrosine kinases and src kinase associated signaling. Src kinases phosphorylate a variety of targets and are direct cellular processes such as endocytosis of receptors, cell proliferation, cell migration metabolism and differentiation. Since Src kinases are implicated in a wide variety of diseases, they have become a target of pharmacological treatment. Inhibitors such as dasatinib (Bristol-Myer Squibb) bind to the active kinase domain and block the function of the Src kinase, Lyn, and the kinase domain of the fusion protein bcr-abl. Other Src kinase/bcr-abl have entered into clinical trials for tumors, including saracatinib, bosutinib, and KX01 (kinex pharmaceuticals). In addition to Src, the Src family contains several other family members: Lyn, Fyn, Yes, Fgr, Hck, Lck, and Bck. The family members share the same kinase domain but each have their own unique domain. Each family member has its own cell type distribution. Src kinases Lyn, Hck, and Fyn are distributed in immune cell types and regulate the function of inflammatory responses by T cells, B cells, and macrophages.
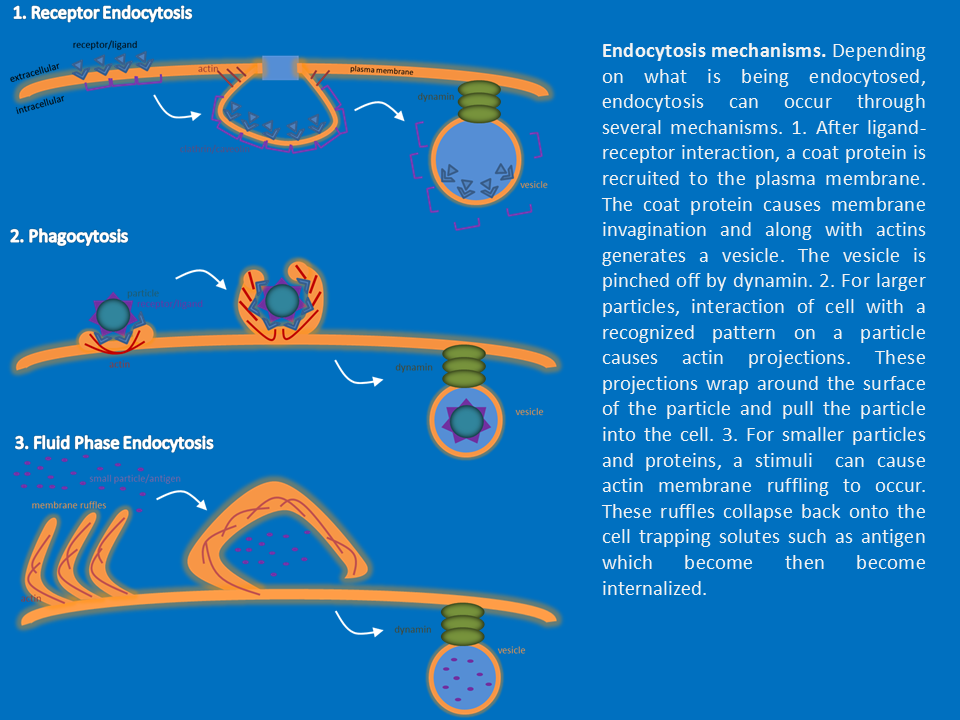
My main involved studying the involvement of src kinases (Src, Lyn, and Hck), WASp and dynamin in macrophage endocytosis. I developed a variety of assays to quantify internalization by each process and determine the contribution of cytoskeletal proteins. I used fluorescence microscopy to visualize the architecture of the vesicles produced and the cytoskeleton structure of macrophages during inflammation. These techniques were used on both cell culture cells and cell’s isolated from different tissues. In addition to endocytosis, I studied the cytokine response by macrophages and the vesicle system the cytokines utilize.function and endocytosis. Along with dendritic cells, macrophages are the front line of defense and detection of pathogens such as bacteria and viruses. In addition, macrophages remove debris such as particles of bone, inhaled dust, and foreign materials. For pathogens, macrophages internalize whole bacteria, whole virus particles, vira/bacterial proteins, and antibody opsinized particles. Macrophages internalize these particles and process the antigens for presentation. The route of internalization differs depending on the context of the antigen. During the course of the project, I obtained experience in techniques necessary to study macrophage internalization of antigens by receptor mediated endocytosis, phagocytosis, and fluid phase endocytosis. Other projects I was involved in included studying the endocytosis of glucose receptors and its effects on glucose metabolism. Another project examined the endocytosis of the EGF receptor and its effects on migration and invasion of cancer cells.
After the completion of the project, I decided to return to virology and joined the laboratory of Dr. William J. Muller MD/Phd in the Department of Pediatrics at Northwestern University. The laboratory focuses on the Human Herpes Virus (HSV-1 and HSV-2) replication and pathogenesis. In conjunction with the laboratory of Dr. Richard Longnecker PhD, the group studies the dependence of HSV-1 and HSV-2 on nectin-1 and HVEM for entry and replication. While nectin-1 is indispensable for infection, little is known why HSV binds HVEM. HSV-1 and HSV-2 infect human epithelial cells and spread to neurons. HSV infects epithelial cells at various sites including the eye, mouth, skin, and genital tract. After establishing a foci of infection, the virus spreads downward through the epithelial layer to peripheral nerves. Once peripheral nerves become infected, the virus travels to the nervous system and establishes latency in a nerve ganglion. The virus sporadically reactivates to travel back to the skin to pass to other individuals. In addition to studying entry, Dr. Muller studies the pathogenesis of HSV-1 and HSV-2 in the neonatal model of infection. Pediatric cases of HSV-1 and HSV-2 differ from adult cases in the severity of the disease. While adults control the infection well, infants up to the age of 6 months on a case-to-case basis develop encephalitis resulting in death and neurologic morbidity.
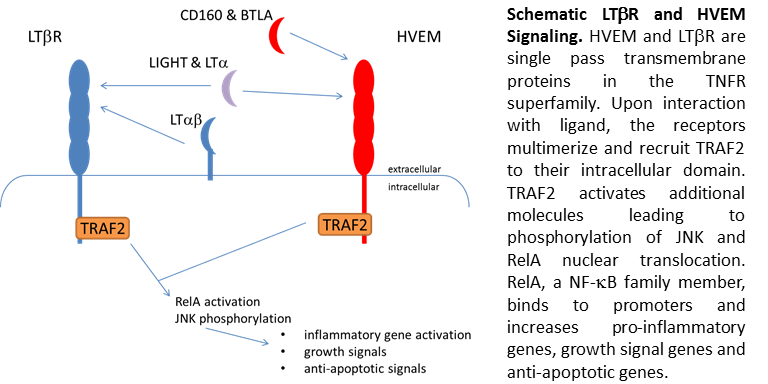
Since the virus binds and utilizes a immune receptor for entry, there is the potential the virus modules inflammation through this interaction to enhance viral replication. Since little was known about the function of HVEM, I decided to investigate NF-kB and JNK signaling through HVEM. HVEM belongs to the tumor necrosis receptor superfamily. Similar to TNF, the other family members and HVEM regulate inflammation, apoptosis and cell survival. HVEM has several activating ligands including lymphotoxin, CD160, BTLA, and LIGHT. Like many other receptors HVEM shares ligands with other receptors, specifically LTbR. Single or co-expression of these ligands on cell surface, leads to different signaling intensities in the presence of ligand. The sharing of ligands allows cells to fine tune signaling from LIGHT and other ligands depending on the cell type. For HVEM, I demonstrated co-expression with LTbR lead to dampening of signals involved in apoptosis and cell survival. These data would suggest HVEM expression may aide in cell survival after LIGHT exposure. These studies contribute to our knowledge of HVEM function and may eventual lead to an understanding of why HSV binds HVEM during entry.
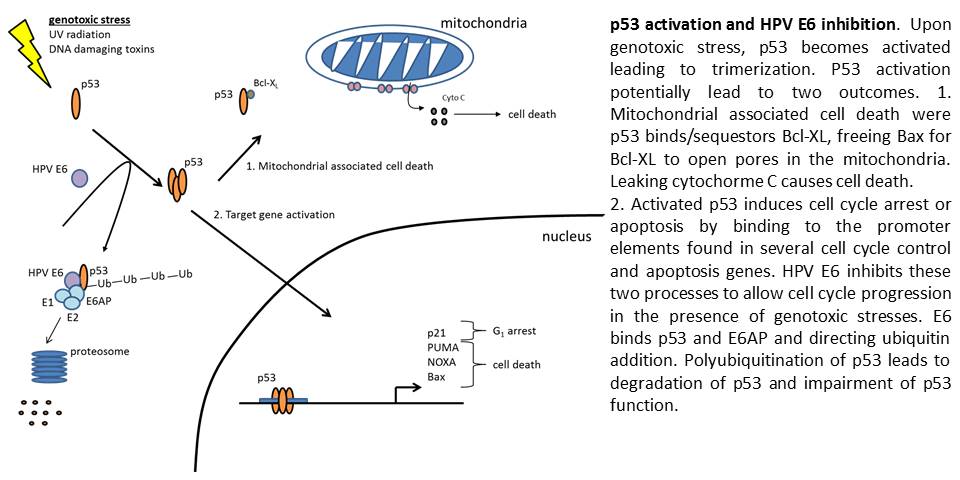
After leaving the Muller lab, I joined the Spiotto lab in Radiation Oncology Department at the University of Chicago. The Spiotto lab focuses on Head and Neck Cancers and HPV oncogenesis. Recently, the incidence of HPV associated cancers has spiked indicating a need for understanding HPV carcinogenesis. While the vaccinated younger population remains protected, the aging unvaccinated population remains at high risk. The lab focuses on the two main tumor oncogenes, E6 and E7. These two oncogenes inactivate p53 and Rb respectively, greatly increasing the chances for tumor development. E6’s inactivation of p53 greatly impairs the ability of the cell to initiate apoptosis following DNA damage and limit pre-cancerous cell development. The fact p53 mutations are found in 50% of cancers highlights the importance of p53 in containing cancer. In addition, the lab studies how other common cancer mutations synergize with HPV oncogenes to lead to cancer development. The overall goal of these studies is to target these oncogenes and tailor treatments for patients.
MICHIGAN STATE UNIVERSITY
MIC 408 ADVANCED MICROBIOLOGY LAB (W)
MIC 463 MEDICAL MICROBIOLOGY
MIC 301 INTRODUCTORY MICROBIOLOGY
MIC 431 MICROBIAL GENETICS
MIC 451 IMMUNOLOGY
MIC 302 INTRO MICROBIOLOGY LAB
MIC 409 EUKARYOTIC CELL BIOLOGY
MIC 413 VIROLOGY
MIC 461 MOLECULAR PATHOGENESIS
MIC 491 CURRENT TOPICS IN MICROBIOLOGY
BCH 461 BIOCHEMISTRY I
BCH 462 BIOCHEMISTRY II
CEM 351 ORGANIC CHEMISTRY I
CEM 352 ORGANIC CHEMISTRY II
CEM 355 ORGANIC LABORATORY I
ANT 316 GENERAL HUMAN ANATOMY
LBS 126 PERSONAL COMPUTERS & NETWORKS
LBS 144 BIOLOGY I: ORGANISMAL BIOLOGY
LBS 145 BIOLOGY II CELL & MOLEC BIO
LBS 165 INTRO CHEMISTRY & PHYSICS I
LBS 165L INTRO CHEMISTRY LAB I
LBS 164 INTRO PHYSICS & CHEMISTRY I
LBS 266 INTRO CHEMISTRY & PHYSICS II
LBS 267 INTRO PHYSICS & CHEMISTRY II
LBS 267L INTRO PHYSICS LAB II 1
LBS 266L INTRO CHEMISTRY LAB II 1
LBS 118 CALCULUS I
LBS 119 CALCULUS II
LBS 164L INTRO PHYSICS LAB I
LBS 331 LITERATURE AND SCIENCE
LBS 133 INTRO SCIENCE & TECH STUDIES
HST 425 AMER/EURO HLTH CARE SINCE 1800
SPN 201 SECOND YEAR SPANISH I
SPN 320 GRAMMAR AND COMPOSITION (W) 3 3.0
SPN 102 ELEMENTARY SPANISH II
ISS 210 SOCIETY AND THE INDIVIDUAL (D)
SPN 202 SECOND-YEAR SPANISH II
ISS 335 NTL DIVERSITY & CHANGE US (N)
IAH 201 U.S. & THE WORLD (D)
LOYOLA UNIVERSITY
BICH 401 MOLECULAR & CELL BIOCHEMISTRY
MIIM 413 BASIC CONCEPTS IMMUNOLOGY
MIIM 414 VIROLOGY
MBIO 417 MOLECULAR BIOLOGY
MIIM 431 MOLECULAR BIOLOGY OF ANIMAL VIRUSES
MBIO 527 SIGNAL TRANSDUCTION
MIIM 411 BASIC MOLECULAR MICROBIOLGY
MBIO 471 COMPREHENSIVE MOLECULAR GENETICS
BMSC 402 STATISTICAL METHDS FOR BIOMEDICAL SCIENCES
BMSC 405 ETHICS IN BIOMEDICAL SCIENCES